Understanding Water Interaction Leads to the Discovery of a New Class of Reversible USP7 Inhibitors that Suppress Tumor Growth
Summary
The Journal of Medicinal Chemistry recently published Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Anti-Tumor Activity. This paper was authored by researchers at RAPT Therapeutics with contributions from Schrödinger’s Christopher Higgs, a Senior Director in the Application Science group who applied FEP+ to help guide RAPT’s chemistry efforts.
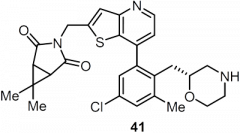
USP7 is a promising target for cancer therapy as its inhibition is expected to decrease function of oncogenes, increase tumor suppressor function, and enhance immune function. The paper used a structure-based drug design strategy to discover a new class of reversible USP7 inhibitors that is highly potent in biochemical and cellular assays and extremely selective for USP7 over other deubiquitinases (DUBs). Their research led to the discovery of compound 41 (Table 1), a highly potent, selective, and orally bioavailable USP7 inhibitor that demonstrated tumor growth inhibition in both p53 wildtype and p53 mutant cancer cell lines, showing that USP7 inhibitors can suppress tumor growth through multiple different pathways.
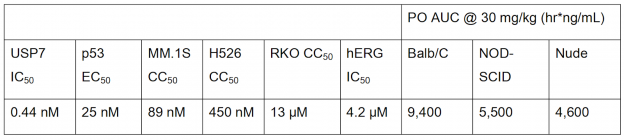
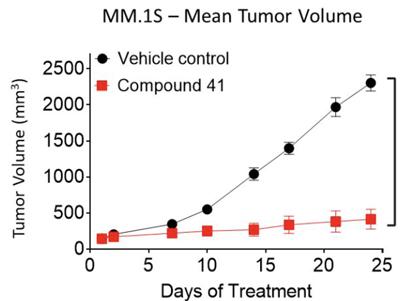
The ability of compound 41 to suppress tumor growth was assessed in an xenograft model using mice. When given a 50 mg/kg oral dose of compound 41 twice daily, nearly complete tumor growth inhibition was observed (Figure 2).
Dr. Higgs explains the paper’s key findings:
Explain what this paper means. Why is it so exciting?
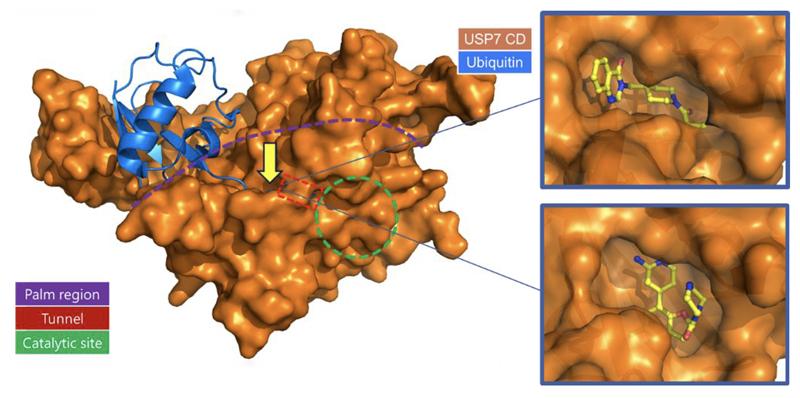
A growing number of proteins involved in different types of cancers are believed to be binding partners or substrates of USP7. This makes USP7 an attractive target for new therapies against several cancers, including ovarian, colon, and neuroblastoma. Hit compounds, identified by high throughput screening or derived from starting points reported in the literature, were co-crystalized with USP7 and found to bind to the allosteric pocket in the “palm” domain. Binding in this pocket is believed to stabilize an inactive conformation of USP7, preventing USP7 catalytic activity. With this data, our structure-based methods, including WaterMap and FEP+, were used to help guide and prioritize compounds to further optimize these series.
The paper showcases the use of the Schrödinger platform to aid drug discovery. Could you explain how Schrödinger software was used in this research?
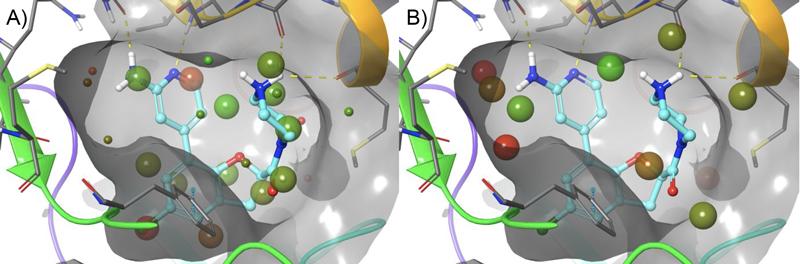
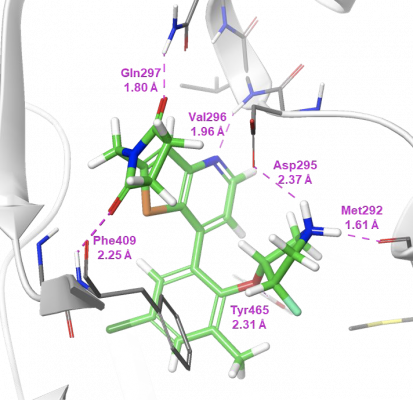
The co-crystal structure of compound 7 in complex with USP7 (Figure 3) was prepared for modeling using the Protein Preparation Wizard in Maestro. Schrödinger’s WaterMap allows for the a 3D visualization of hydration sites in the binding pocket, which provides an understanding of each site’s entropy, enthalpy, and free energy, thus illuminating how different compounds could interact with waters in the binding pocket and predict their relative stability. Using this method in combination with FEP+, Schrödinger scientists were able to provide critical information for helping determine which compounds were likely to be strong binders.
How does this process help you design compounds with optimal molecular properties?
Initial studies using WaterMap identified a small cluster of unfavorable hydration sites , adjacent to the aminopyridine, that with the appropriate functional group, could be displaced to gain further potency (Figure 4). A talented team of Medicinal Chemists at RAPT designed compounds to try and take advantage of this region of the pocket, while FEP+ was used to help prioritize their synthesis queue. This approach allows for fewer DMTA (design-make-test-analyze) cycles while allowing more chemical space to be explored. In addition, FEP+ was used to explore other parts of the molecule, not covered in this publication, which also enabled improvements in potency and PK.
This publication is significant for cancer research, is that right?
USP7 is a very promising target for additional cancer treatments due to the large number of interactions it appears to mediate. However, more work needs to be done to fully characterize USP7’s role in these signaling pathways. This paper’s goal is to aid in that effort.
How did this work between RAPT Therapeutics and Schrödinger come about?
Schrödinger’s Applications Science group works closely with users of our software around the world, providing expert guidance on best practices and training on new functionality Schrödinger’s computational platform. The group does, on occasion, get actively involved in discovery projects under modeling services agreements, particularly where a new application area is being explored or to show the value our platform can bring in a real world context.