FEP Protocol Builder
Spend more time on your science and less time fine-tuning protocol settings for challenging systems

Spend more time on your science and less time fine-tuning protocol settings for challenging systems
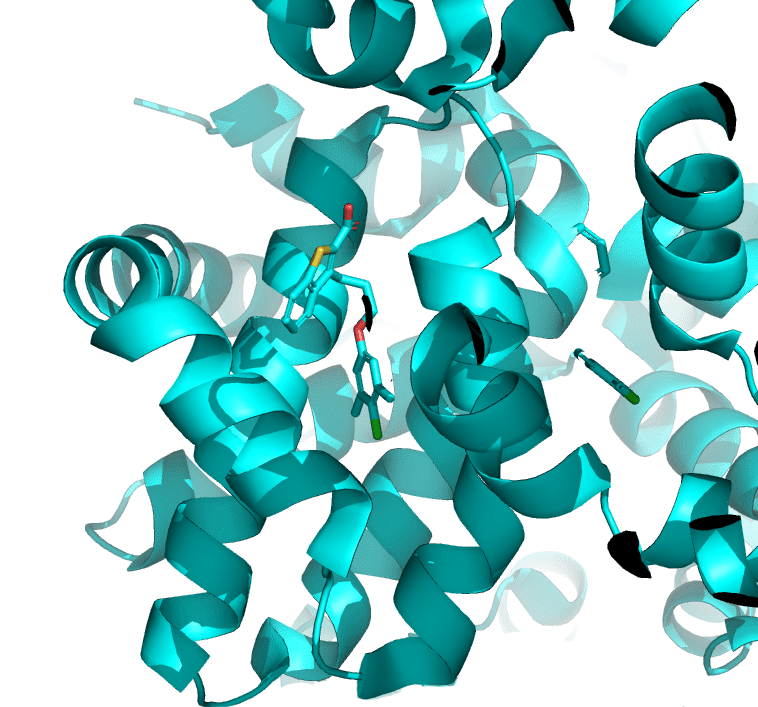
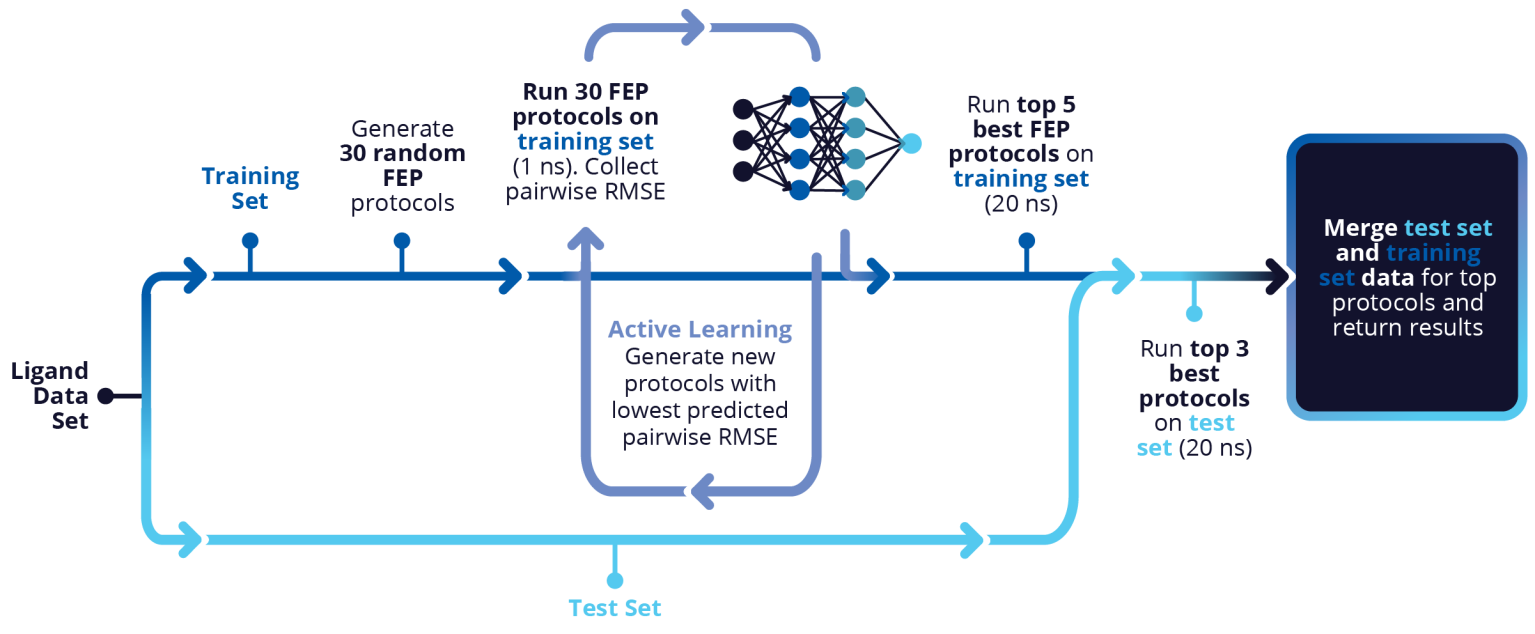
FEP Protocol Builder is an automated machine learning workflow for FEP+ model optimization. This workflow is designed for systems with insufficient predictive accuracy using default settings or after initial manual protocol optimization attempts. FEP Protocol Builder saves researcher time and increases the chances of successfully enabling FEP+ by efficiently identifying an optimized predictive model for your system of interest.
| Protocol | ASN879 rotamer | ASP892 charge state | Water Model | Reference ligands | REST Residues | Pairwise RMSE (kcal/mol) |
|---|---|---|---|---|---|---|
| Default | Flipped | ASP | SPC | 4887, 4897 | – | 2.48 |
| FEP-PB | Not Flipped | ASH | TIP4PD | 4898, 4901 | A:LEU 772 A:PHE 893 | 1.07 |
Fully automated FEP Protocol Builder generated optimized FEP protocol with improved pairwise RMSE compared to default model. All protocols were performed with the same 15 congeneric ligands with modifications at 2 R-groups with known affinity data from PDB ID: 2IVV. FEP Protocol Builder was applied to quickly and rigorously validate the amenability of the target to the Schrödinger platform.
de Oliveira C, et al. J. Chem. Inf. Model. 2023, 63, 17, 5592–5603.
Abel R, et al. Acc. Chem. Res. 2017, 50, 7, 1625–1632.