FEP+
High-performance free energy calculations for drug discovery

High-performance free energy calculations for drug discovery
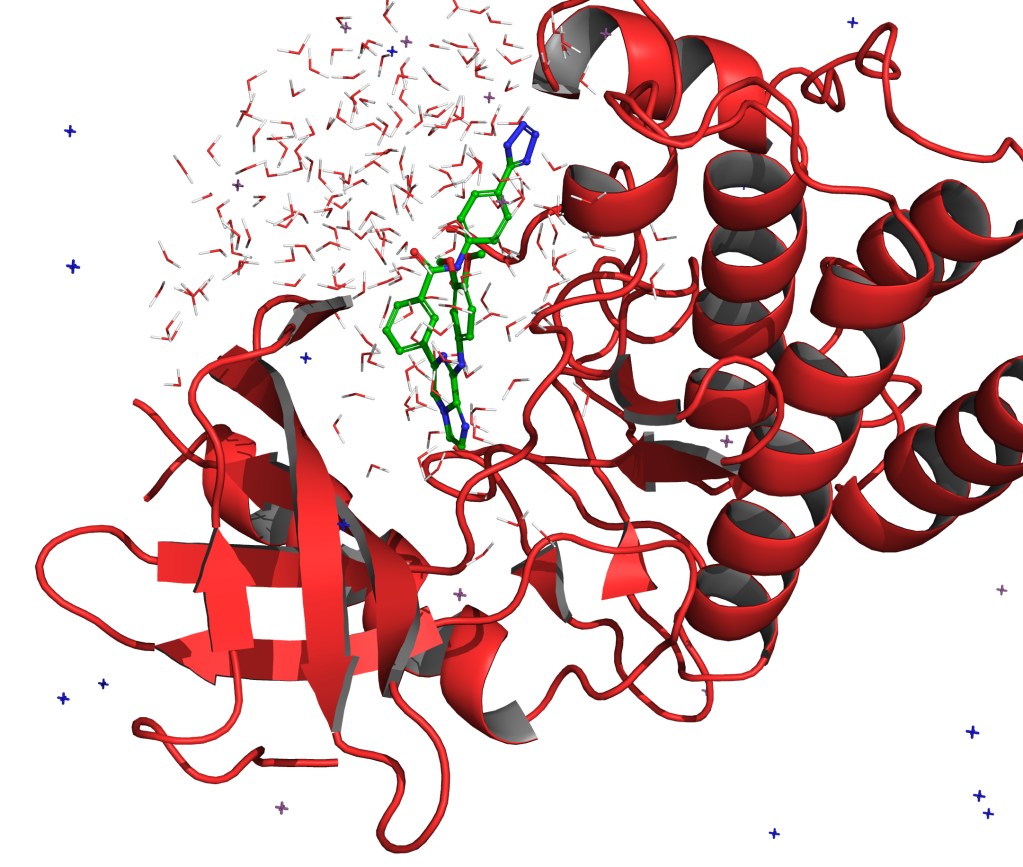
FEP+ is Schrödinger’s proprietary, physics-based free energy perturbation technology for computationally predicting protein-ligand binding at an accuracy matching experimental methods, across broad chemical space.
Leverage FEP+ as an accurate in silico binding affinity assay to drive rapid virtual design cycles and focus experimental efforts on only the highest quality ideas
Optimize multiple properties simultaneously, including potency, selectivity, and solubility, to improve the profile and developability of small and large molecules
Synthesize novel and challenging chemistry with a high degree of confidence through prospective application of FEP+

Predictive accuracy approaching experiment (1 kcal/mol) as demonstrated in large-scale validation studies across diverse ligands and protein classes
Widely adopted by leading pharma and biotech companies, with several drug candidates in the clinic driven by FEP+
Supports the broadest range of applications and perturbation types common in drug discovery scenarios and consistently expanded through active R&D
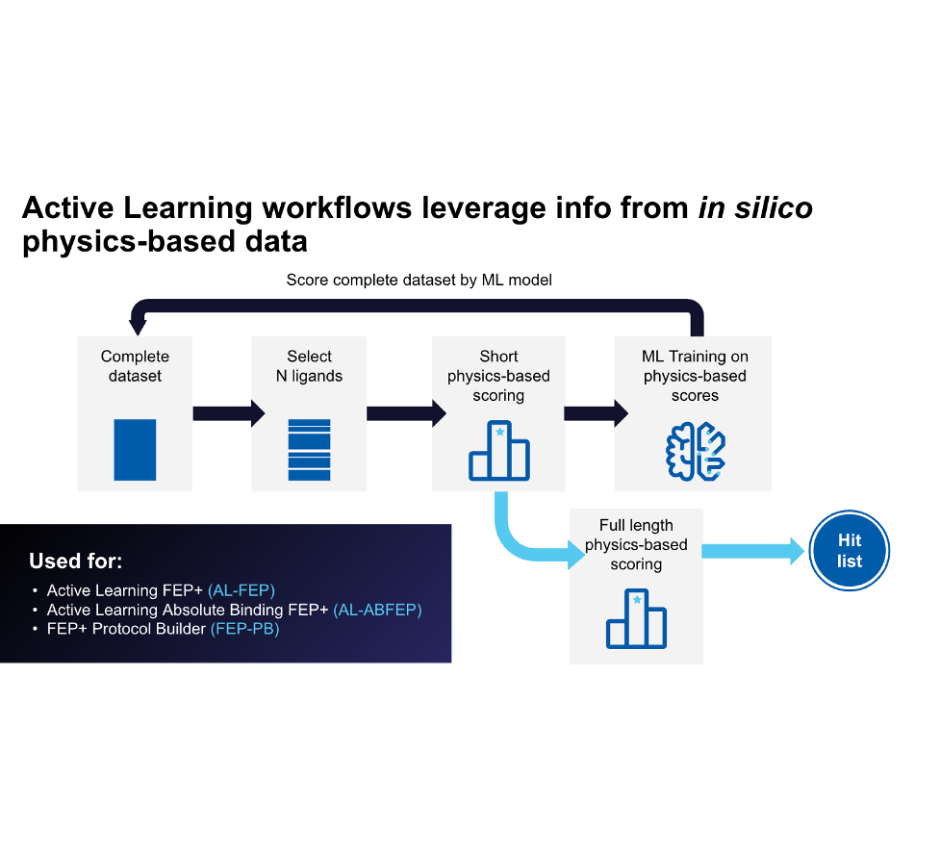
Leverage a well-validated, automated workflow which trains a machine learning model on project-specific FEP+ data to allow processing of up to millions of compounds with highly accurate FEP+ calculations efficiently.

Level-up your FEP+ skills and enroll in our online molecular modeling course, Free Energy Calculations for Drug Design with FEP+.
View CourseDiscover how Schrödinger’s technology is being used to solve real-world research challenges.
Digital chemistry platform provides scale and accuracy to drive high precision molecular design
Collaborative enterprise platform and physics-based digital assays empower a team of experts to tackle a challenging target
Single-edge FEP+ integrated in LiveDesign

Schrödinger has a strategic partnership with NVIDIA to optimize our computational drug discovery platform for NVIDIA GPU technology.

Learn more about the related computational technologies available to progress your research projects.
Fully-integrated, cloud-based design system for ultra-large scale chemical space exploration and refinement
A modern, comprehensive force field for accurate molecular simulations
Accurate ligand binding mode prediction for novel chemical matter, for on-targets and off-targets
Browse the list of peer-reviewed publications using Schrödinger technology in related application areas.
Level up your skill set with hands-on, online molecular modeling courses. These self-paced courses cover a range of scientific topics and include access to Schrödinger software and support.
Learn how to deploy the technology and best practices of Schrödinger software for your project success. Find training resources, tutorials, quick start guides, videos, and more.