Prime
A powerful and innovative solution for accurate protein structure prediction

A powerful and innovative solution for accurate protein structure prediction
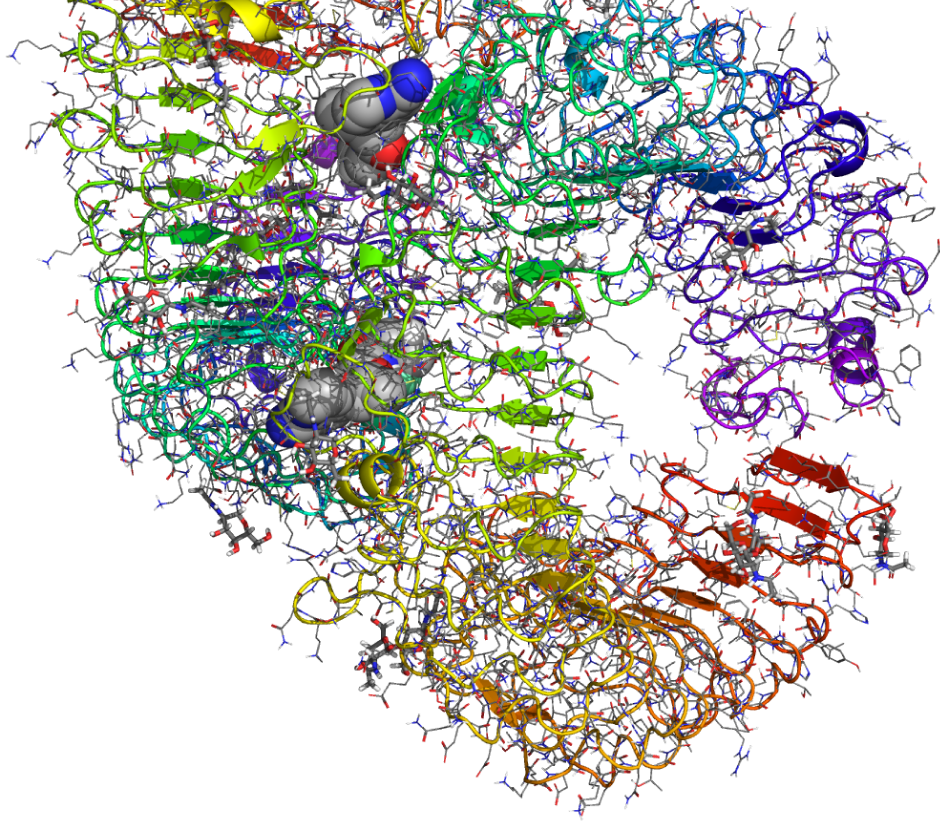
Prime is a fully-integrated protein structure prediction solution that incorporates homology modeling and fold recognition into a single solution. Prime includes an intuitive step-by-step interface that takes users through the workflow of structure prediction by supplying helpful default settings for each stage of the process.
Discover how Schrödinger technology is being used to solve real-world research challenges.
Get answers to common questions and learn best practices for using Schrödinger’s software.

Learn more about the related computational technologies available to progress your research projects.
Accurate ligand binding mode prediction for novel chemical matter, for on-targets and off-targets
Physics-based solution for rapid and accurate prediction of passive membrane permeability
Browse the list of peer-reviewed publications using Schrödinger technology in related application areas.
Level up your skill set with hands-on, online molecular modeling courses. These self-paced courses cover a range of scientific topics and include access to Schrödinger software and support.
Learn how to deploy the technology and best practices of Schrödinger software for your project success. Find training resources, tutorials, quick start guides, videos, and more.