SiteMap
Fast, accurate, and intuitive binding site identification

Fast, accurate, and intuitive binding site identification
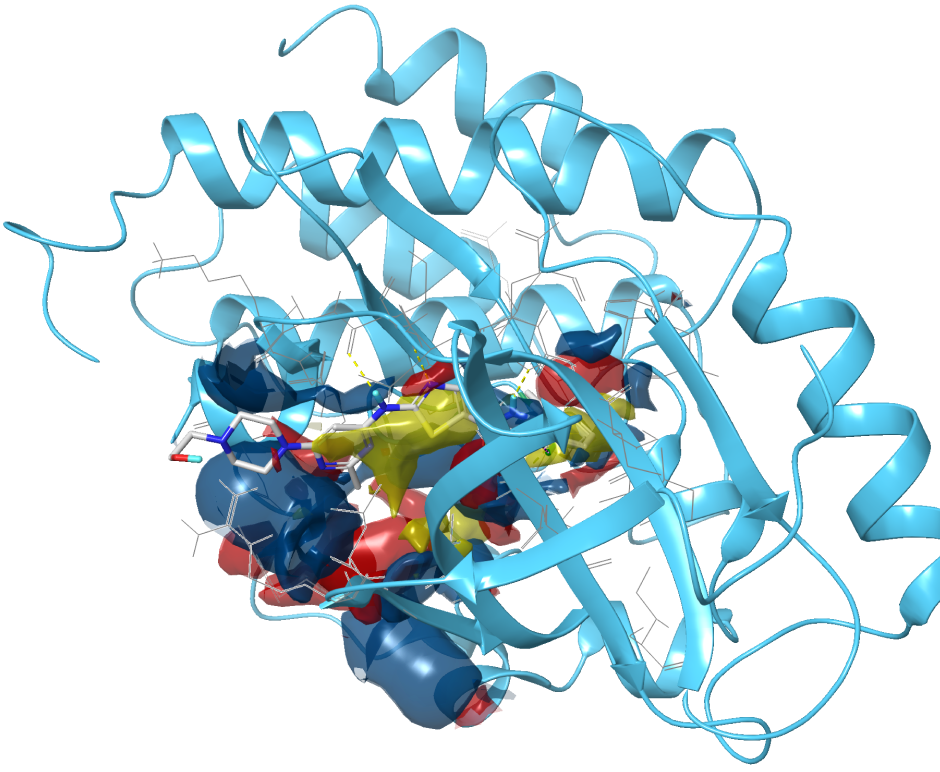
Identifying druggable pockets is an important early challenge in a structure-based first-in-class drug discovery project. However, locating these sites in drug design projects is often challenging. SiteMap’s proven algorithm helps identify binding sites, including allosteric binding sites and protein-protein interfaces, and evaluate their druggability. In addition to impacting lead discovery, SiteMap can assist researchers in lead optimization by providing insights into potential ligand-receptor interactions which can then guide modification of lead compounds to increase their binding potency.
Locate binding sites whose size, functionality, and extent of solvent exposure meet user specifications. Rank possible binding sites using physics-based criteria tuned to eliminate those not likely to be pharmaceutically relevant using SiteScore, the scoring function used to assess a site’s propensity for ligand binding.
Easily visualize binding sitesIdentify regions within the binding site suitable for occupancy by hydrophobic groups or by ligand hydrogen-bond donors, acceptors, or metal-binding functionality. Distinguish different binding site sub-regions which allows for ready assessment of a ligand’s complementarity.
Easily use identified sites to set up docking models for structure-based virtual screening experiments with Glide.
Use generated binding site maps to guide what types of ligand modifications would be expected to promote binding.
Get answers to common questions and learn best practices for using Schrödinger’s software.

Learn more about the related computational technologies available to progress your research projects.
State-of-the-art, structure-based method for assessing the energetics of water solvating ligand binding sites for ligand optimization
Browse the list of peer-reviewed publications using Schrödinger technology in related application areas.
Level up your skill set with hands-on, online molecular modeling courses. These self-paced courses cover a range of scientific topics and include access to Schrödinger software and support.
Learn how to deploy the technology and best practices of Schrödinger software for your project success. Find training resources, tutorials, quick start guides, videos, and more.