Design of a highly selective, allosteric, picomolar TYK2 inhibitor using novel FEP+ strategies
Computationally-guided, structure-based drug design strategy drives discovery of potentially best-in-class TYK2 inhibitor
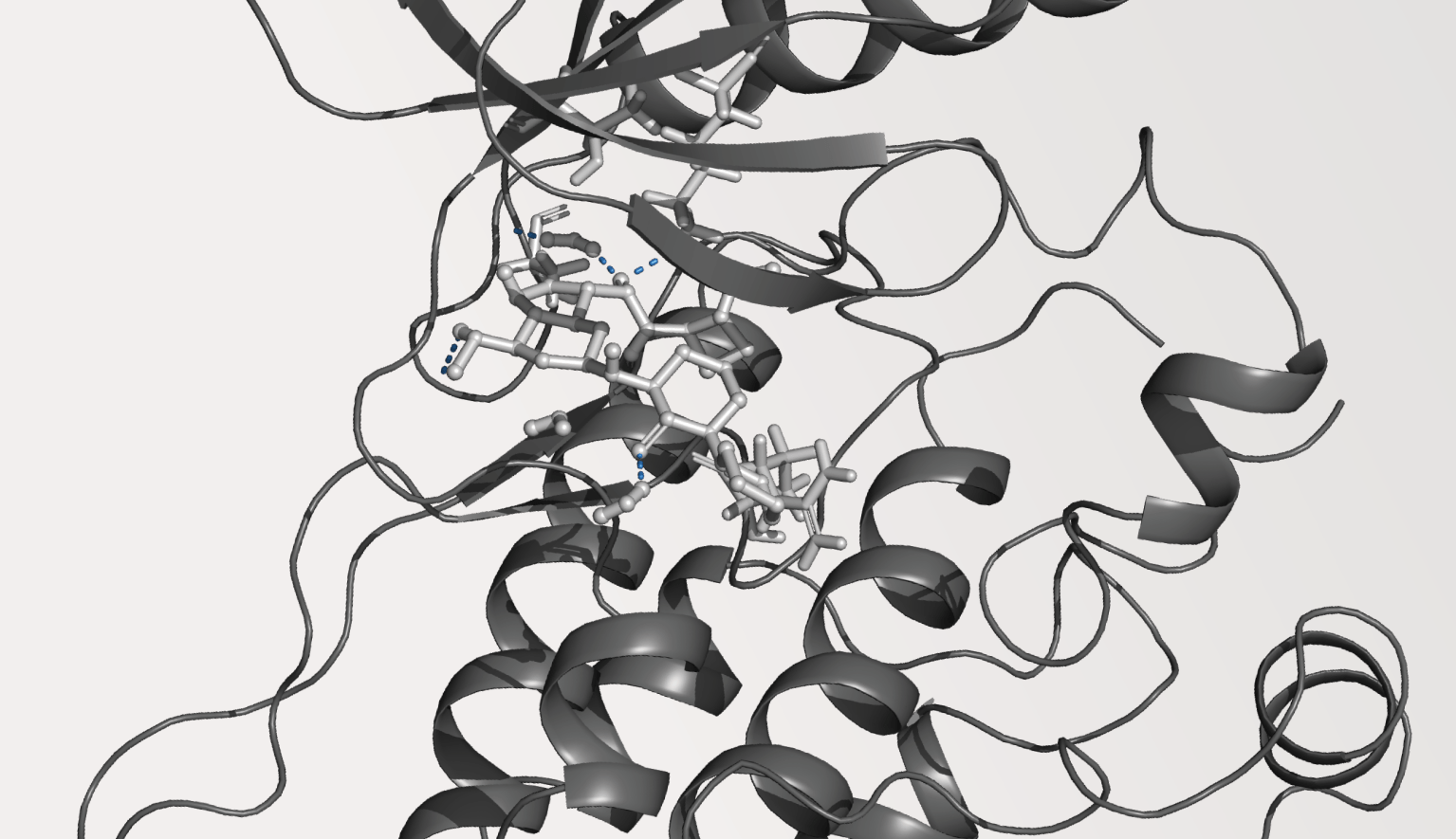
TYK2 kinase
Collaborative program, small molecule
Nimbus Therapeutics
Inflammatory diseases, psoriasis
Phase 2b → Phase 3 clinical trial
“Our teams designed, iterated, and optimized leads using next-generation, physics-based technologies, which allowed us to rapidly refine our discovery methodologies. This partnership provided Nimbus with unparalleled computational horsepower and unprecedented FEP+ modeling efforts.”
— Craig Masse
Former Head of Medicinal Chemistry,
Nimbus Therapeutics
Design challenge
JAK/TYK kinases are key signaling molecules in a variety of inflammatory diseases. In psoriasis, for example, inflammation is driven by increased interferon and cytokine release, which is regulated by these JAK/TYK kinases. To date, marketed drugs have targeted the JH1 kinase domain of these multi-domain protein complexes. However, these approved JAK inhibitor drugs have known safety issues related to heart function, clotting, and thrombosis due to non-selective inhibition of other JAK kinases (JAK1, JAK2, JAK3).
Scientists from Nimbus Therapeutics began working on this challenge with the goal of developing a potent and highly selective TYK2 inhibitor that improved clinical activity while avoiding off-target side effects. In 2016, Nimbus and Schrödinger scientists teamed up on the project, first starting by targeting the catalytic JH1 kinase domain and later pivoting to the regulatory domain JH2 based on the release of promising data in the literature.
Targeting the TYK2 JH1 catalytic domain: Breakthroughs in TYK2 selectivity and solubility using large-scale FEP+
The high sequence similarity of TYK2 within the JAK family posed a significant selectivity challenge — the TYK2, JAK1, JAK2, and JAK3 ligand binding sites are practically identical. To design a highly selective and potent molecule that maintains activity while avoiding off-target liabilities, the team used large-scale, physics-based simulations to differentiate the binding between the kinases. First, free energy perturbation calculations using FEP+ were used to predict the potency on-target on TYK2, as well as to predict the potency off-target on JAK2 and JAK3.1 Next, the team introduced a novel approach for predicting aqueous solubility using absolute FEP+ in order to address ADME properties during modeling.2
With these modeling approaches in hand, the next turning point in the program was the development of a large-scale FEP+ scoring campaign to tackle this multiparameter optimization (MPO) challenge. The team leveraged extensive medicinal chemistry and modeling expertise to enumerate 4,000 high-quality idea molecules, all of which had passed preliminary triage through MM-GBSA and physical property filters.3 Then FEP+ was used to score the potency, selectivity, and solubility of the molecules (Figure 1). Accomplishing this large scale application of FEP+ scoring required deploying FEP+ to cloud computing resources at scale, which since this initial effort has become a commonly used approach. Ultimately, 46 molecules were prioritized on the basis of the FEP+ calculations for synthesis, and 9 of these molecules were found to meet potency, selectivity, and solubility criteria of the project in later experimental testing. This resulted in a development candidate targeting the TYK2 JH1 domain that was highly selective against JAK2 and moderately selective against JAK1 and JAK3.

Pivot to TYK2 JH2 allosteric domain enabled by FEP+ de novo core design and virtual screening
Around the same time, new research from scientists at Bristol Myers Squibb confirmed that highly selective TYK2 inhibitors could be achieved by targeting the JH2 allosteric site.4 The project team made the important decision to pivot — changing their focus to developing a TYK2 inhibitor targeting the JH2 regulatory domain. This biological validation along with the deep structural enablement available for TYK2 and the JAK family member pseudokinase domains provided the team with an opportunity to engage in a multi-pronged, structure-based design strategy.
FEP+ was used to guide compound design strategies with de novo core exploration resulting in the identification of the pyrrazolono-pyridine and pyridino-imidazole cores, which displayed promising potency and domain selectivity but were hindered by several ADME liabilities (Figure 2).5 A large-scale structure-based virtual screen enabled screening of 2.4M commercially available compounds against an ensemble of receptor models based on known JH2 crystal structures from the PDB using Glide followed by WScore — resulting in three structurally distinct hit classes (Figure 2).5 Each hit displayed some promising level of JH2 affinity, but the pyrazolo-pyrimidine core was chosen for further exploration based on its steric and electronic similarity to the imidazopyridazine core published by BMS. These structure-based design efforts allowed the team to identify multiple picomolar JH2 scaffolds from these hits in a period of 3 months.
In order to progress these hits, the team again employed an MPO paradigm with large scale modeling, this time in two stages. First, quick modeling of physicochemical properties, as well as passive permeability, was performed to ensure the compounds progressed to resource intensive FEP+ models and synthesis would stay in a reasonable chemical space. Second, compounds were put through rigorous FEP+ potency predictions — targeting picomolar potency. All models and MPOs were collected in LiveDesign, Schrödinger’s cloud-based enterprise informatics platform, within different LiveReports for different design objectives which the team would review together. Using this MPO strategy, the team carried out parallel ligand FEP+ modeling against TYK2 JH2 as well as other JAK family kinase domains in order to design analogs with high JH2 affinity and JAK family domain selectivity.
Within two years of pivoting to the allosteric TYK2 program, the team had declared a development candidate. NDI-034858 is a picomolar inhibitor tuned precisely to target the JH2 domain of TYK2, as shown by its exquisitely clean selectivity profile (Figure 3).
In November 2022, Nimbus reported positive topline results for once-daily, oral dosing with NDI-034858 in Phase 2b clinical trials.6 Furthermore, by February 2023 Takeda had acquired the program.7 Now referred to as TAK-279, this highly selective, oral TKY2 inhibitor has best-in-class potential among allosteric inhibitors for treatment of psoriasis, as well as multiple other immune-mediated diseases.8

Enabling digital technologies to drive discovery programs
FEP+
High-performance free energy calculations for large-scale prediction of potency, selectivity, and solubility
LiveDesign
Collaborative enterprise informatics platform for centralizing access to virtual and wet lab project data and powerful computational predictions
References
-
Advancing drug discovery through enhanced free energy calculations.
Abel et al. Acc. Chem. Res. 2017, 50, 7, 1625–1632.
-
Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Leit et al. Bioorg. Med. Chem. Lett. 2022 Oct 1, 73,128891.
-
Accelerating drug discovery through tight integration of expert molecular design and predictive scoring.
Abel et al. Curr. Opin. Struct. Biol. 2017 Apr, 43, 38-44.
-
Tyrosine kinase 2-mediated signal transduction in T lymphocytes Is blocked by pharmacological stabilization of its pseudokinase domain.
Tokarski et al. J Biol Chem. 2015 Apr 24, 290(17), 11061-74; Identification of imidazo[1,2-b]pyridazine TYK2 pseudokinase ligands as potent and selective allosteric inhibitors of TYK2 signalling. Moslin et al. MedChemComm. 2016 Dec 15, 8(4), 700-712.
-
Discovery of a potent and selective tyrosine kinase 2 inhibitor: TAK-279.
Leit et al. J. Med. Chem. 2023, 66, 15, 10473–10496.
-
Nimbus Therapeutics announces positive topline results for phase 2b clinical trial of allosteric TYK2 inhibitor in psoriasis.
Nimbus Therapeutics. 2022.
-
Takeda completes acquisition of Nimbus Therapeutics’ TYK2 program subsidiary.
Takeda. 2023.
-
Takeda announces positive results in phase 2b study of investigational TAK-279, an oral, once-daily TYK2 inhibitor, in people with moderate-to-severe plaque psoriasis.
Takeda. 2023.
Software and services to meet your organizational needs
Industry-Leading Software Platform
Deploy digital drug discovery workflows using a comprehensive and user-friendly platform for molecular modeling, design, and collaboration.
Modeling Services
Leverage Schrödinger’s team of expert computational scientists to advance your projects through key stages in the drug discovery process.
Scientific and Technical Support
Access expert support, educational materials, and training resources designed for both novice and experienced users.