FEP+ for Biologics
High-performance free energy calculations for biologics discovery

High-performance free energy calculations for biologics discovery
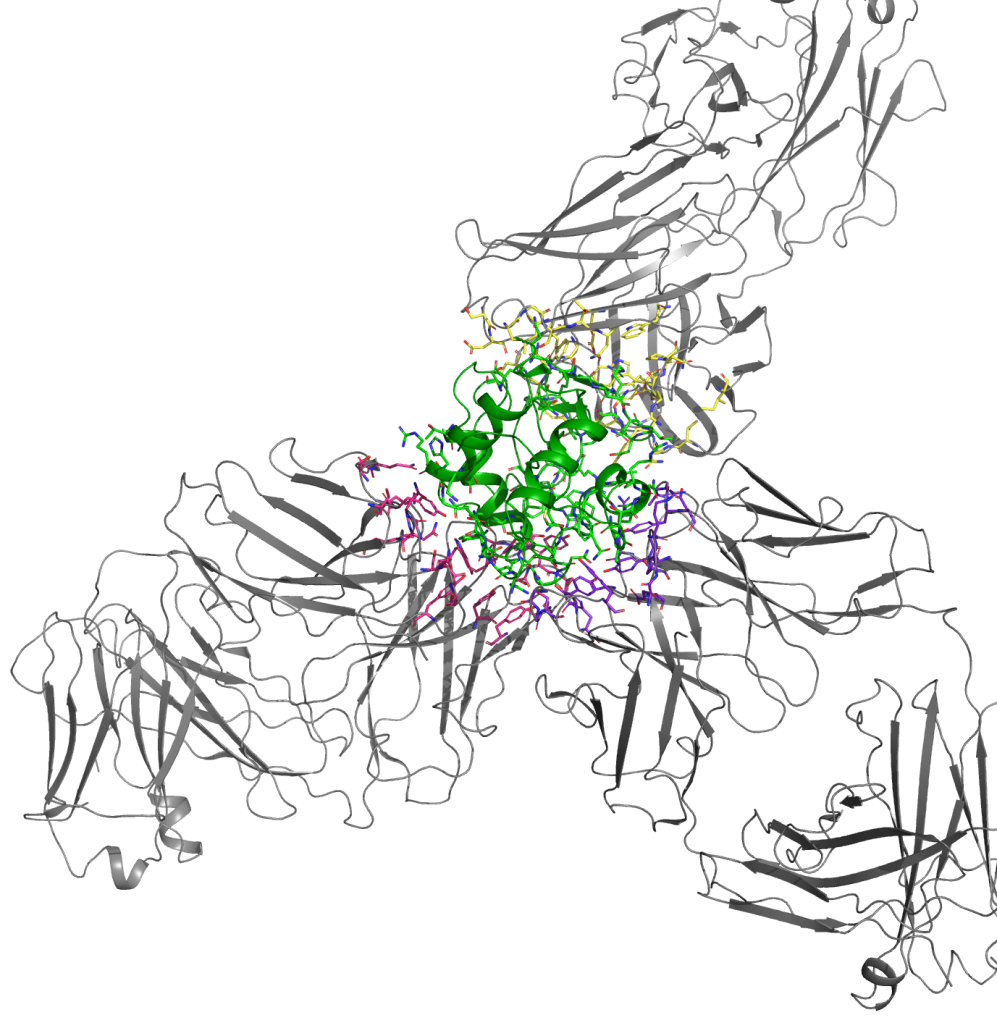
FEP+ is Schrödinger’s proprietary, physics-based free energy perturbation technology. Use FEP+ in your biologics discovery projects to computationally predict protein mutation effects at an accuracy matching experimental methods to within ~1 kcal/mol.
Enhance or decrease binding affinity of a protein to its target
Tune preferential binding of a protein to one target over another
Extending breadth of binding across multiple targets
Engineer pH-dependent association or dissociation between two proteins
Enhance physical stability of a protein-based biologic
Optimize properties like solubility, chemical stability, and physical stability

Accurately quantify the effects of mutations on protein stability and protein-protein binding affinities during the optimization of antibodies, antigens, peptides, enzymes, and other biologic products.
Rapidly model single mutations on multiple sites simultaneously to achieve consistent and reliable prediction of mutational effects and dramatically improve the efficiency of FEP+ calculations.
Our workflow leverages FEP+ Residue Scan and Protein FEP+ technologies to enable precise and efficient exploration of protein mutations to simultaneously optimize stability, binding affinity, specificity, pH-dependent binding, and cross-reactivity.
Correlation plots between MM/GBSA, FEP+ Residue Scan and Protein FEP+ calculations (y-axis), and relative experimental affinity measurements (x-axis), shown in kcal/mol. All calculations performed on the same system, mutations to and from proline currently excluded from FEP+ Residue Scan results. Pearson correlation coefficient (R2) shown at top left of each plot.
Access refined workflows across multiple biological modalities
Sampson, et al. J Mol Biol. 2024, 436, 16.
Scarabelli G, et al. J Mol Biol. 2022, 434, 2.
Katz D, et al. Mar. Drugs 2021, 19(7), 367.
Lihan M, et al. Prot Sci. 2023, 32, 2, e4557.
Haufe Y, et al. ACS Chem Neurosci. 2023, 14, 24, 4311–4322.

If you’re new to using FEP+, get a refresher on the fundamental concepts of relative binding free energy perturbation (FEP) calculations.
Get answers to common questions and learn best practices for using Schrödinger’s software.
Learn more about the related computational technologies available to progress your research projects.
High-performance molecular dynamics (MD) engine providing high scalability, throughput, and scientific accuracy
A modern, comprehensive force field for accurate molecular simulations
Deploy digital drug discovery workflows using a comprehensive and user-friendly platform for molecular modeling, design, and collaboration.
Leverage Schrödinger’s computational expertise and technology at scale to advance your projects through key stages in the drug discovery process.
Access expert support, educational materials, and training resources designed for both novice and experienced users.