Ligand Designer
Intuitive, interactive 3D ligand design for hit-to-lead and lead optimization

Intuitive, interactive 3D ligand design for hit-to-lead and lead optimization
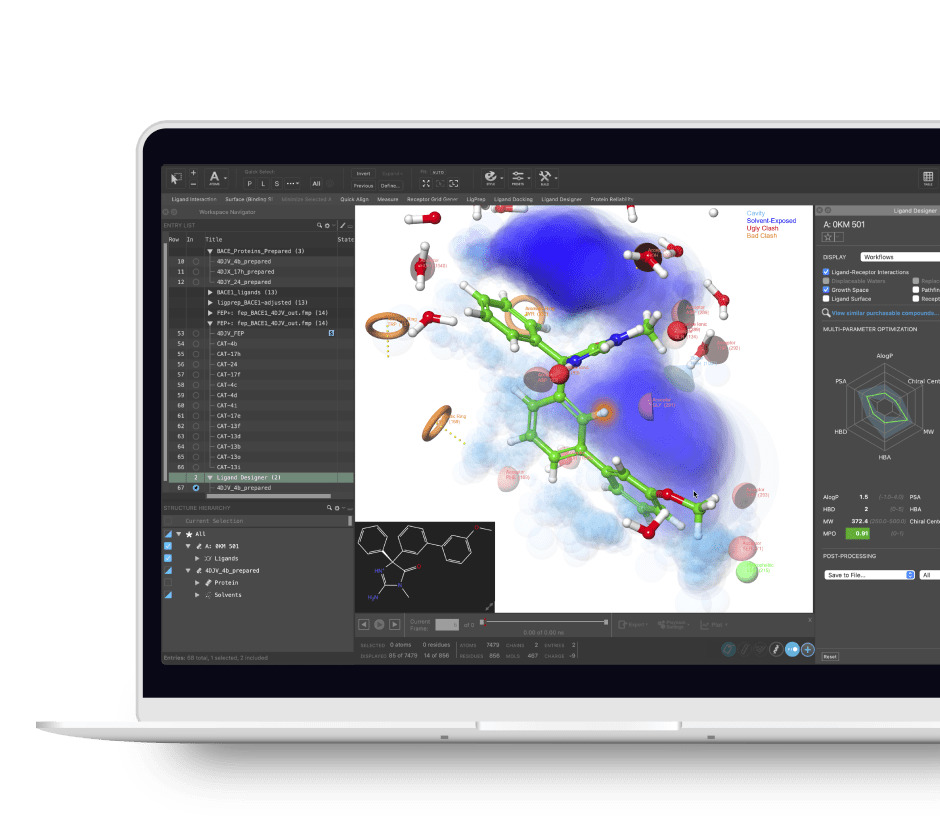
Ligand Designer provides powerful, yet easy to use 3D visualization and ligand building capability that is scientifically proven while making it fun to design ligand modifications in 2D or 3D and see how those changes are likely to impact protein-ligand complex structures. Cutting through the complexity of visualizing 3D information, the Ligand Designer uses concepts from Augmented Reality to enable you to layer relevant information including a novel grow space to quickly recognize where ligand modifications are most desirable.
Ligand Designer democratizes the 3D design process by putting the power of common medicinal chemistry tactics at your fingertips. The tool is available in Maestro and LiveDesign.

Choose your own adventure with manual or guided design to evaluate ideas. Common medicinal chemistry workflows are included so you can test ideas automatically and avoid information overload.
Design in 2D and see the structures in the workspace; in 2½ D and see the representation of the binding pocket with key information; or design in 3D with full control.
Easily assess the impact of ligand changes by visualizing the complex geometry while reducing the noise of un-needed information to find the next idea. Controls are pre-programmed to fit your function.
Tailor it to your chemistry of interest by using synthetically tractable enumeration or R-group enumeration.
Glide docking, automated ideation using R-group libraries or Pathfinder, Prime’s Macrocycle, BREED and WaterMap are all used in Ligand Designer.
Rationalize SAR, drive potency and tune selectivity using WaterMap to compute the entropy and enthalpy of “hydration sites.”

Learn more about the related computational technologies available to progress your research projects.
State-of-the-art, structure-based method for assessing the energetics of water solvating ligand binding sites for ligand optimization
An easy-to-use pharmacophore modeling solution for ligand- and structure-based drug design
Level up your skill set with hands-on, online molecular modeling courses. These self-paced courses cover a range of scientific topics and include access to Schrödinger software and support.
Learn how to deploy the technology and best practices of Schrödinger software for your project success. Find training resources, tutorials, quick start guides, videos, and more.