- Documentation
Epik
Accurately and rapidly predict the aqueous phase pKa values and protonation state distributions of complex, drug-like molecules.
- Publication
- Feb 13, 2025
Towards automated physics-based absolute drug residence time predictions
Smith Z, et al. ChemRxiv, 2025, Preprint
- Publication
- Jan 30, 2025
Accurate physics-based prediction of binding affinities of RNA- and DNA-targeting ligands
Abramyan, et al. J. Chem. Inf. Model. , 2025, 65, 3, 1392–1403
- Publication
- Apr 23, 2024
FEP augmentation as a means to solve data paucity problems for machine learning in chemical biology
Burger PB, et al. J. Chem. Inf. Model., 2024, 64(9), 3812–3825
- Publication
- Apr 6, 2023
Epik: pKa and Protonation State Prediction through Machine Learning
Ryne C. Johnston, et al. Journal of Chemical Theory and Computation, 2023
- Publication
- Nov 2, 2021
The transcriptional corepressor CtBP2 serves as a metabolite sensor orchestrating hepatic glucose and lipid homeostasis
Sekiya, et al. Nat Commun, 2021, 12(1), 6315
- Publication
- Mar 4, 2017
Adverse Drug Reactions Triggered by’the Common HLA-B*57:01 Variant: A Molecular Docking Study
Van Den Driessche, et al. J. Cheminform., 2017, 9 (13), 1-17
- Publication
- Sep 28, 2014
Discovery of Thienoquinolone Derivatives as Selective and ATP Non-Competitive CDK5/p25 Inhibitors by Structure-Based Virtual Screening
Chatterjee, et al. Bioorg. Med. Chem., 2014, 22, 6409-6421
- Publication
- Jun 19, 2013
Boosting virtual screening enrichments with data fusion: Coalescing hits from two-dimensional fingerprints, shape, and docking
Sastry, et al. J. Chem. Inf. Model., 2013, 53, 1531-1542
- Publication
- May 24, 2012
Testing physical models of passive membrane permeation
Leung, et al. J. Chem. Inf. Model., 2012, 52(6), 1621-1636
- Publication
- Mar 6, 2012
Predicting and improving the membrane permeability of peptidic small molecules
Rafi, et al. J. Med. Chem., 2012, 55(7), 3163-3169
Events
 Event
Materials Science
Event
Materials Science
- Feb 4, 2026
3rd Industrial Polymers & CPG Summit 2026
We are pleased to invite you to the 3rd Industrial Polymers & CPG Summit. Mark your calendars for February 3-4, 2026, in the picturesque city of Heidelberg.
 Event
Life Science
Event
Life Science
- Feb 5, 2026
Diverse computational strategies enable the discovery of p38α-MK2 molecular glues
In this webinar, Schrödinger’s medicinal and computational chemists will show how they used a multipronged computational design strategy to discover multiple structurally diverse, potent, and highly selective molecular glues.
 Webinar
Materials Science
Webinar
Materials Science
- Feb 10, 2026
Physics-driven ML to accelerate the design of layered multicomponent electronic devices
Schrödinger has developed a machine learning (ML) framework that enables users to predict key performance metrics of multilayered electronic devices from simple, intuitive descriptions of their architecture and operating conditions.
Webinars
 Webinar
Life Science
Webinar
Life Science
- Feb 11, 2026
Introducing RetroSynth: Breaking the synthesis bottleneck with AI and physics-based modeling
Join us for the introduction of RetroSynth, Schrödinger’s AI-driven synthesis planning platform. RetroSynth is engineered to accelerate and scale conventional retrosynthesis by harnessing advanced deep learning algorithms.
 Webinar
Life Science
Webinar
Life Science
- Feb 12, 2026
From silos to synthesis: Fostering collaborative AI through platform integration with LiveDesign ML
Successfully leveraging AI investments demands a platform that delivers unparalleled predictive accuracy and seamless operationalization.
 Webinar
Life Science
Webinar
Life Science
- Feb 18, 2026
Schrödinger デジタル創薬セミナー: Into the Clinic ~計算化学がもたらす創薬プロセスの変貌~ 第22回
Predictive toxicology solutions: Actionable, structure-based insights to dial-out tox liabilities early
Documentation
- Documentation
Learning Path: Oligonucleotide Modeling
A structured overview of tools and workflows for nucleic acids in drug discovery.
- Documentation
WaterMap
Efficiently converged MD simulations are run with explicit water molecules, and resultant trajectories are analyzed to cluster hydration sites.
- Documentation
SiteMap
Identify binding sites, including allosteric binding sites and protein-protein interfaces, and evaluate their druggability.
Tutorials
- Tutorial
Structure-Based Virtual Screening using Glide
Prepare receptor grids for docking, dock molecules and examine the docked poses.
- Tutorial
Ligand Binding Pose Generation for FEP+
Generate starting poses for FEP simulations for a series of BACE1 inhibitors using core constrained docking.
- Tutorial
Homology Modeling of Protein-Ligand Binding Sites with IFD-MD
Create a homology model of TYK2 from JAK3 and including a bound ligand. Compare this model with the crystal structure for TYK2 bound to 4GIH.
Training Videos
 Video
Life Science
Video
Life Science
Getting Going with Maestro BioLuminate
A free video series introducing the basics of using Maestro Bioluminate.
 Video
Life Science
Video
Life Science
- Video
Introducing Ligand Designer
An overview of the LigandDesigner workflow, Editing in 2D and 3D, using display options and overlays, and accessing the Admin Panel.
Publications
- Publication
- Oct 13, 2025
Accelerated in silico discovery of SGR-1505: A potent MALT1 allosteric inhibitor for the treatment of mature B-cell malignancies
Nie Z, et al. J. Med. Chem., 2025
- Publication
- Oct 12, 2025
Discovery of highly potent noncovalent inhibitors of SARS-CoV-2 main protease through computer-aided drug design
Okabe A, et al. J Med Chem, 2025
- Publication
- Sep 17, 2025
Accurate hydration free energy calculations for diverse organic molecules with a machine learning force field
Xie, et al. ChemRxiv, 2025, 1, Preprint
Case Studies
 Case Study
Life Science
Materials Science
Case Study
Life Science
Materials Science
 Case Study
Life Science
Case Study
Life Science
 Case Study
Life Science
Case Study
Life Science
White Papers
 White Paper
Life Science
White Paper
Life Science
- Jan 29, 2026
FEP+ Pose Builder — maximizing utility and productivity in FEP simulations
FEP+ Pose Builder is a methodological advancement introduced as an integrated feature to drastically enhance accessibility, user-friendliness, and productivity within the FEP+ pipeline.
 White Paper
Life Science
White Paper
Life Science
- Oct 29, 2024
20 Years of Glide: A Legacy of Docking Innovation and the Next Frontier with Glide WS
Glide has long set the gold standard for commercial molecular docking software due to its robust performance in both binding mode prediction and empirical scoring tasks, ease of use, and tight integration with Schrödinger’s Maestro interface and molecular discovery workflows.
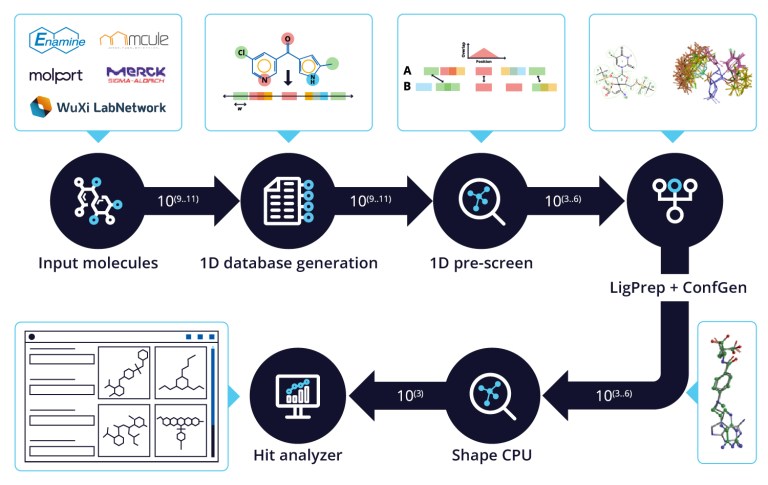 White Paper
Life Science
White Paper
Life Science
Quick Reference Sheets
- Quick Reference Sheet
GlideMap
A one-page guide to using the GlideMap GUI for ligand placement guided by experimental density.
- Quick Reference Sheet
Synthesis Queue LiveReport
Use Freeform columns to track the status of compounds in a synthesis queue.
- Quick Reference Sheet
Modeling Queue LiveReport
Learn how to use Freeform columns and an Auto-Update Search to create compound progression workflows.

Latest insights from Extrapolations blog
Training & Resources
Online certification courses
Level up your skill set with hands-on, online molecular modeling courses. These self-paced courses cover a range of scientific topics and include access to Schrödinger software and support.
Free learning resources
Learn how to deploy the technology and best practices of Schrödinger software for your project success. Find training resources, tutorials, quick start guides, videos, and more.