- Tutorial
Introduction to T Cell Receptor Modeling with BioLuminate
Structure preparation, visualization and analysis of key interactions in the TCR-peptide-MHC complex.
- Tutorial
Protein pKa Prediction with Constant pH Molecular Dynamics
Determine pKa values and protonation states for protein residues.
- Tutorial
Improving the Thermostability of T4 Lysozyme Using Protein FEP+ Guided Design
Increase protein thermostability by filling a buried cavity through mutation with protein FEP+.
- Tutorial
Computational Ellipsometry
Learn how to compute the refractive index and extinction coefficient of systems of organic optoelectronics.
- Tutorial
Refining crystallographic protein-ligand structures using GlideXtal and Phenix/OPLS
Re-dock and refine ligand pose in a crystal structure with GlideXtal.
- Tutorial
Understanding and Visualizing Target Flexibility
Evaluate PDB temperature factors, align binding sites, and use MD to identify flexibility.
- Tutorial
Re-scoring Docked Ligands with MM-GBSA
Optimize binding poses and re-score results of a small virtual screen.
- Tutorial
Approximating Protein Flexibility without Molecular Dynamics
Soften potentials in Glide and run induced-fit docking for side chain conformational changes and loop refinement.
- Tutorial
Introduction to MD Trajectory Analysis with Desmond
Analyze an all-atom Desmond MD trajectory to study protein-ligand interactions.
- Tutorial
FEP Solubility
Perform a Free Energy of Perturbation (FEP) Solubility simulation on ibuprofen.
- Tutorial
Disulfide Bond Engineering
Run cysteine scanning to identify residues that could be mutated to cysteine to improve thermal stability and facilitate crystallization.
Events
 Event
Materials Science
Event
Materials Science
- Jan 28th-30th, 2026
【1月28日(水)~30日(金)】nano tech 2026 出展
展示ブースでは、セミナー開催のほか、先進の技術について専門技術者が解説し、お客様からのご質問にお答えします。 小間番号:3W-H07 出展ゾーン:【材料・素材】マテリアルゾーン
 Webinar
Life Science
Webinar
Life Science
- Jan 29, 2026
From silos to synthesis: Fostering collaborative AI through platform integration with LiveDesign ML
Successfully leveraging AI investments demands a platform that delivers unparalleled predictive accuracy and seamless operationalization.
 Event
Materials Science
Event
Materials Science
- Feb 3rd-4th, 2026
3rd Industrial Polymers & CPG Summit 2026
We are pleased to invite you to the 3rd Industrial Polymers & CPG Summit. Mark your calendars for February 3-4, 2026, in the picturesque city of Heidelberg.
Webinars
 Webinar
Life Science
Webinar
Life Science
- Jan 29, 2026
From silos to synthesis: Fostering collaborative AI through platform integration with LiveDesign ML
Successfully leveraging AI investments demands a platform that delivers unparalleled predictive accuracy and seamless operationalization.
 Webinar
Life Science
Webinar
Life Science
- Feb 11, 2026
Introducing RetroSynth: Breaking the synthesis bottleneck with AI and physics-based modeling
Join us for the introduction of RetroSynth, Schrödinger’s AI-driven synthesis planning platform. RetroSynth is engineered to accelerate and scale conventional retrosynthesis by harnessing advanced deep learning algorithms.
 Webinar
Life Science
Webinar
Life Science
- Feb 18, 2026
Schrödinger デジタル創薬セミナー: Into the Clinic ~計算化学がもたらす創薬プロセスの変貌~ 第22回
Predictive toxicology solutions: Actionable, structure-based insights to dial-out tox liabilities early
Documentation
- Documentation
Learning Path: Oligonucleotide Modeling
A structured overview of tools and workflows for nucleic acids in drug discovery.
- Documentation
WaterMap
Efficiently converged MD simulations are run with explicit water molecules, and resultant trajectories are analyzed to cluster hydration sites.
- Documentation
SiteMap
Identify binding sites, including allosteric binding sites and protein-protein interfaces, and evaluate their druggability.
Tutorials
- Tutorial
Structure-Based Virtual Screening using Glide
Prepare receptor grids for docking, dock molecules and examine the docked poses.
- Tutorial
Ligand Binding Pose Generation for FEP+
Generate starting poses for FEP simulations for a series of BACE1 inhibitors using core constrained docking.
- Tutorial
Antibody Visualization and Modeling in BioLuminate
Visualize, build, and evaluate antibody models, analyze an antibody for various characteristics, dock an antigen to an antibody.
Training Videos
 Video
Life Science
Video
Life Science
Getting Going with Maestro BioLuminate
A free video series introducing the basics of using Maestro Bioluminate.
 Video
Life Science
Video
Life Science
- Video
Introducing Ligand Designer
An overview of the LigandDesigner workflow, Editing in 2D and 3D, using display options and overlays, and accessing the Admin Panel.
Publications
- Publication
- Oct 13, 2025
Accelerated in silico discovery of SGR-1505: A potent MALT1 allosteric inhibitor for the treatment of mature B-cell malignancies
Nie Z, et al. J. Med. Chem., 2025
- Publication
- Oct 12, 2025
Discovery of highly potent noncovalent inhibitors of SARS-CoV-2 main protease through computer-aided drug design
Okabe A, et al. J Med Chem, 2025
- Publication
- Sep 17, 2025
Accurate hydration free energy calculations for diverse organic molecules with a machine learning force field
Xie, et al. ChemRxiv, 2025, 1, Preprint
Case Studies
 Case Study
Life Science
Materials Science
Case Study
Life Science
Materials Science
 Case Study
Life Science
Case Study
Life Science
 Case Study
Life Science
Case Study
Life Science
White Papers
 White Paper
Life Science
White Paper
Life Science
- Oct 29, 2024
20 Years of Glide: A Legacy of Docking Innovation and the Next Frontier with Glide WS
Glide has long set the gold standard for commercial molecular docking software due to its robust performance in both binding mode prediction and empirical scoring tasks, ease of use, and tight integration with Schrödinger’s Maestro interface and molecular discovery workflows.
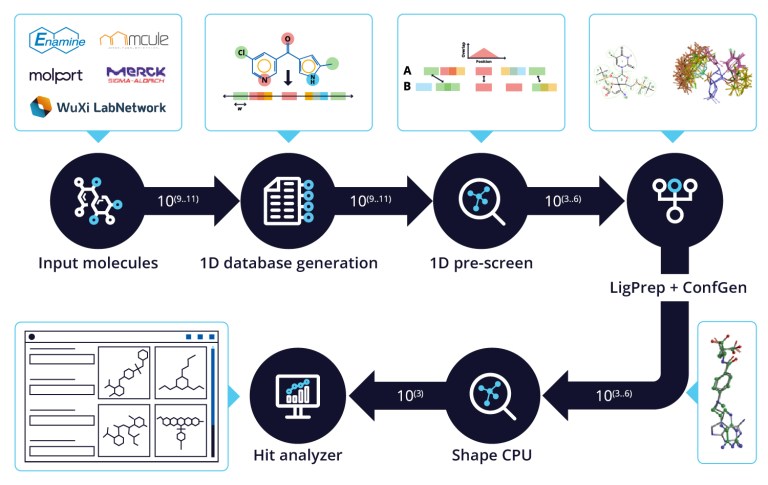 White Paper
Life Science
White Paper
Life Science
 White Paper
Life Science
White Paper
Life Science
Quick Reference Sheets
- Quick Reference Sheet
GlideMap
A one-page guide to using the GlideMap GUI for ligand placement guided by experimental density.
- Quick Reference Sheet
Synthesis Queue LiveReport
Use Freeform columns to track the status of compounds in a synthesis queue.
- Quick Reference Sheet
Modeling Queue LiveReport
Learn how to use Freeform columns and an Auto-Update Search to create compound progression workflows.

Latest insights from Extrapolations blog
Training & Resources
Online certification courses
Level up your skill set with hands-on, online molecular modeling courses. These self-paced courses cover a range of scientific topics and include access to Schrödinger software and support.
Free learning resources
Learn how to deploy the technology and best practices of Schrödinger software for your project success. Find training resources, tutorials, quick start guides, videos, and more.