JUN 25, 2024
Expediting FEP+ model optimization for challenging systems with a fully automated, machine learning-driven workflow
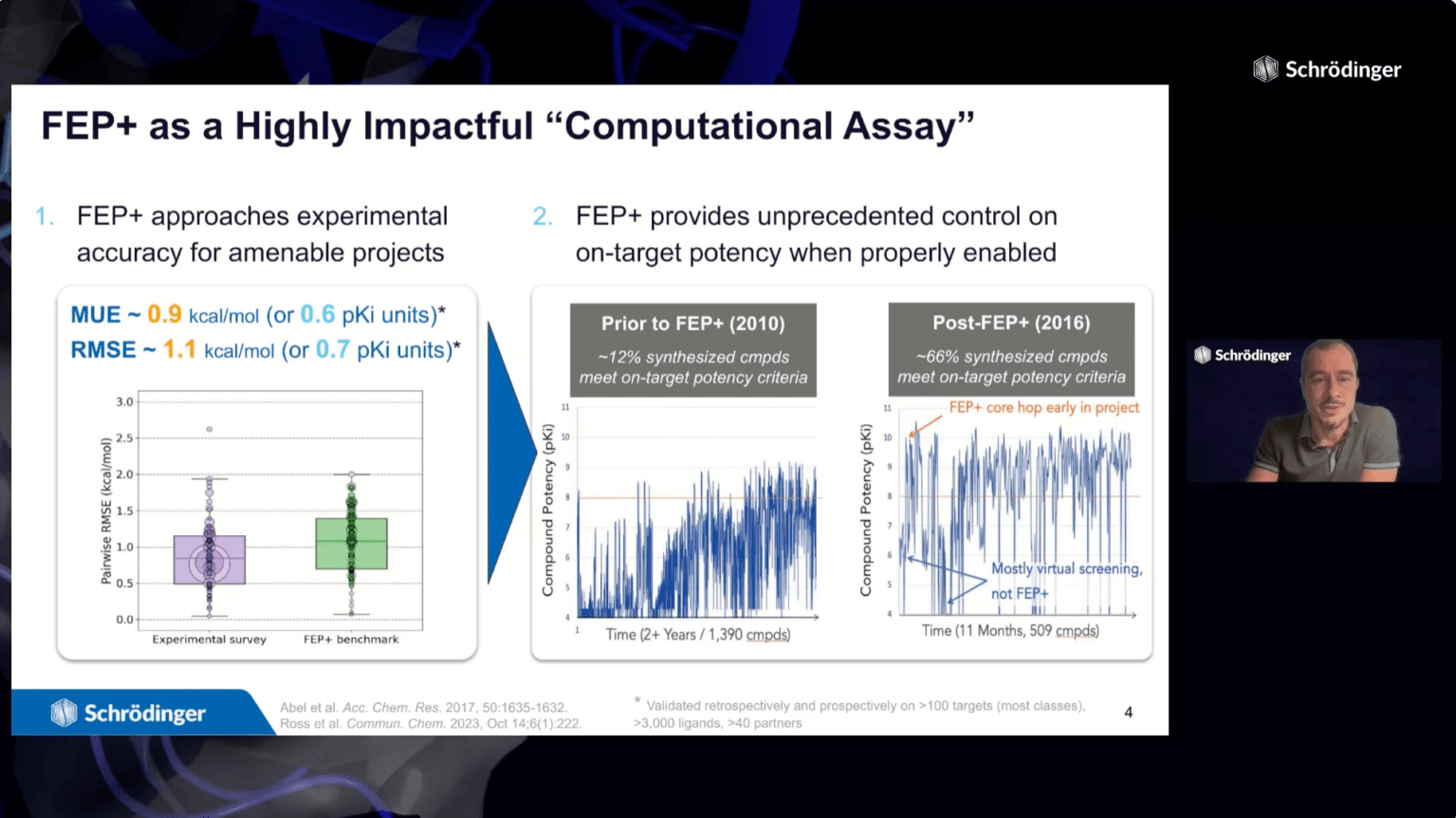
FEP+ is a powerful predictive technology in drug discovery – with applications from hit discovery through lead optimization. A critical first step in deploying FEP+ is to validate and optimize the model for the protein-ligand system of interest. While some systems perform well with out-of-the-box FEP+ settings, others require manual protocol refinement.
In this webinar, we will introduce Schrödinger’s FEP+ Protocol Builder, an automated machine learning workflow for FEP+ model optimization. This workflow is designed for systems with insufficient predictive accuracy using default settings or after initial manual protocol optimization attempts. FEP+ Protocol Builder saves researcher time and increases the chances of successfully enabling FEP+ by efficiently identifying an optimized predictive model for your system of interest.
Join us as we share key features of FEP+ Protocol Builder and highlight case studies that have shown success utilizing the technology to accelerate projects.
Highlights:
- Overview of FEP+ Protocol Builder technology, which uses an Active Learning workflow to iteratively search the protocol parameter space to develop accurate FEP+ protocols
- Case studies on effective prospective use of protocols generated by FEP+ Protocol Builder in drug discovery programs
- Comparison of time efficiency between manual and automated FEP+ optimization
- Details on requirements and services options to leverage the technology in your own program
- Overview of the different options to enable FEP+ at almost all levels of structural information for your protein-ligand system of interest
Our Speakers

Jeremie Vendome
Senior Director, Applications Science, Schrödinger
Dr. Jeremie Vendome, senior director of applications science, joined Schrödinger in 2015. He received his PhD from the Ecole Normale Superieure in France and completed his training at Columbia University in the lab of Prof. Barry Honig, where he worked on various problems related to protein-protein interaction energetics and specificity by combining computational approaches and experiments. Prior to joining Schrödinger, Jeremie acquired 10+ years of drug discovery experience both as a computational chemist in the industry and as the head of a CADD collaborative platform at Columbia Medical Center. At Schrödinger, he has held several roles of increasing responsibility and has continuously been at the interface between the company’s latest technological developments and their applications in active drug discovery projects. Most recently, Jeremie has been spearheading Schrödinger’s research enablement initiative, meant to advance drug discovery programs though key stages by providing access to our latest technologies and workflows at scale as a collaborative service.

Jordan Epstein
Product Manager, Schrödinger
Jordan Epstein joined Schrödinger in 2017 after studying Chemistry at New York University. Upon joining Schrödinger as a software developer, he worked on products such as FEP+, Desmond, and LiveDesign. More recently, he transitioned into his role as product manager where he has helped to see FEP+ Protocol Builder to its full release.

Sathesh Bhat
Executive Director, Therapeutics Group, Schrödinger
Sathesh Bhat, Ph.D., executive director in the therapeutics group, joined Schrödinger in 2011. He is responsible for overseeing computational chemistry efforts on internal and partnered drug discovery programs at Schrödinger. Previously, Sathesh worked at both Merck and Eli Lilly leading computational efforts in several drug discovery programs. He obtained his Ph.D. from McGill University, which involved developing structure-based methods to predict binding free energies. Sathesh has co-authored multiple patents and publications and continues to publish on a wide variety of topics in computational chemistry.