Release Notes
Release 2022-4
Small Molecule Drug Discovery
Platform Environment
Maestro Graphical Interface
- Create and share custom visualization Presets [2022-4]
- Added support to mutate DNA/RNA to standard nucleobases [2022-4]
- New Workflow Action Menu to guide to next steps for Plot Rigid/Relaxed scans [2022-4]
- First full release of the new 2D sketcher [2022-4]
- Get Going with Maestro Video Series added to Documentation [2022-4]
Workflows & Pipelining [KNIME Extensions]
- Includes the latest version of KNIME (v4.6.1) [2022-4]
- The number of matches can now be controlled in the Phase screening node [2022-4]
In LiveDesign:
- When deploying a model the suitable KNIME protocol is chosen automatically and the latest version of the protocol uploaded [2022-4]
- Distribution of calculations is controlled from the model admin page [2022-4]
- Model changes from the LiveDesign Admin page can be preserved when overwriting an existing model [2022-4]
- A new administration node to move, archive and unarchive models [2022-4]
Target Validation & Structure Enablement
Protein Preparation
- Significant speedup when opening the Protein Preparation Workflow interface on Windows [2022-4]
- Reduced verbosity of Protein Preparation Workflow log file by limiting irrelevant CCD bond assignment error notices [2022-4]
- Protein Reliability Report will generate TEST reflections on-the-fly, if not available in provided .cv file, and report RSCC values [2022-4]
- Updated PROPKA to (latest) version 3.4 [2022-4]
Protein X-Ray Refinement
- Introduction of GlideXtal command line tool for automatic ligand fitting in crystallographic electron density maps [2022-4]
- PrimeX minimization is able to use structure factors in CIF format [2022-4]
- In Phenix/OPLS can now remove all entities clashing with crystal mates [2022-4]
- Phenix/OPLS is more robust to missing atoms in standard residues [2022-4]
Cryo-EM Model Refinement
- Beta GlideEM interface for ligand placement into cryo-electron density maps [2022-4]
- GlideEM now accepts gzipped (CCP4, MRC, MAP) files as input [2022-4]
Multiple Sequence Viewer/Editor
- Beta release of Protein Family Alignment and Annotation [2022-4]
- A new category named ‘Family Feature Calculation’ located in the ‘Other Tasks’
- Menu exposes protein family alignment and annotation
- Supports kinase and GPCR Alignments
- Supports annotation of GPCR regions
- Dendrogram hover tooltip to display distance information [2022-4]
IFD-MD
- Membrane-bound IFD-MD tutorial [2022-4]
- Covalent ligand IFD-MD tutorial [2022-4]
FEP+
- Show user-friendly message when undefined stereochemical centers are introduced [2022-4]
- Improved usability of FEP+ group panel to manage protonation and tautomeric states ensemble – for more accurate ΔΔG predictions [2022-4]
Constant pH Simulations (Beta)
- Improved usability of constant pH simulations for protein pKa calculations with friendly outputs [2022-4]
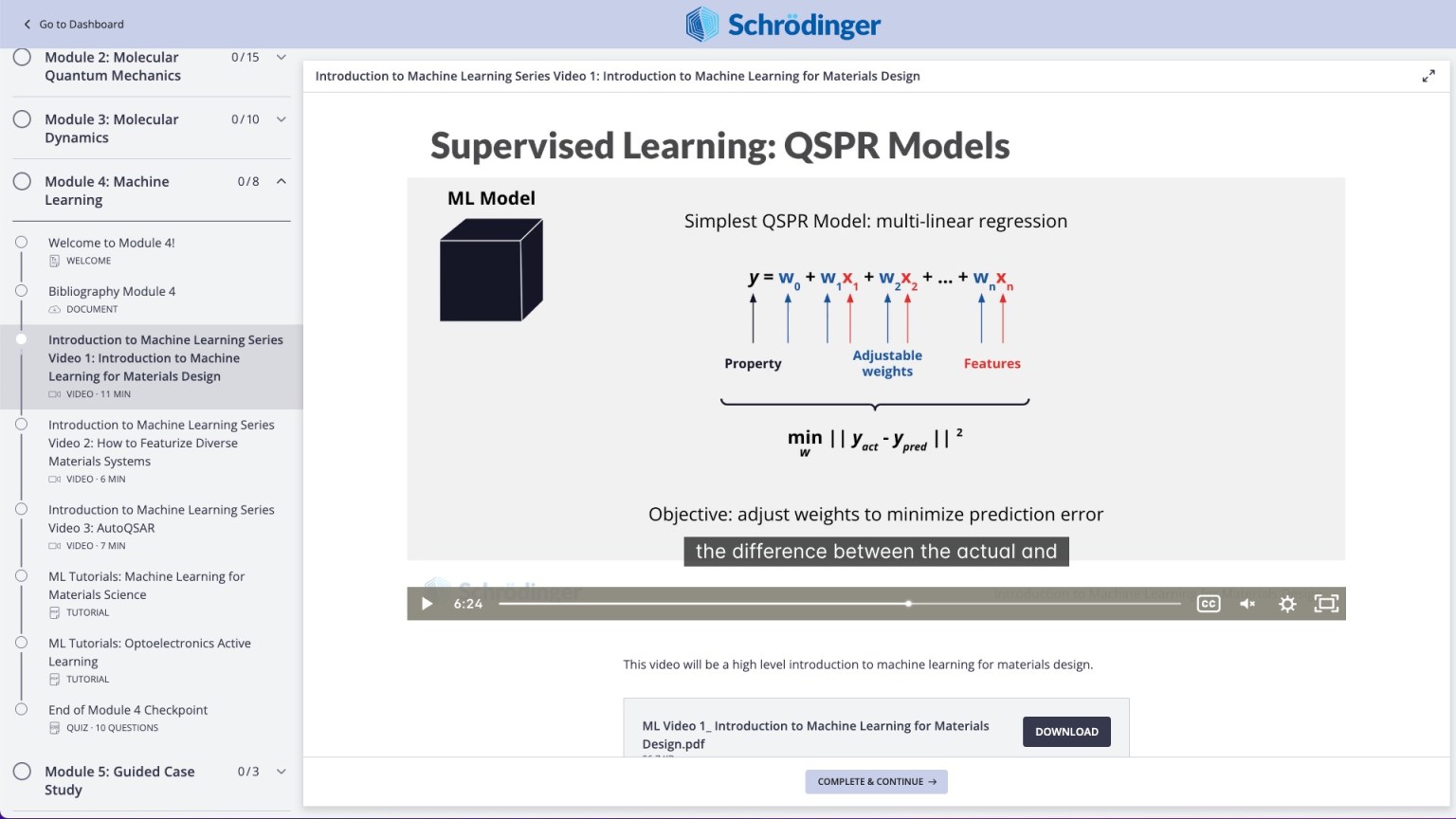
AutoQSAR
- Added MACCs keys for ligand featurization in DeepAutoQSAR [2022-4]
- Include ElasticNetCV model (strongly l1/l2 regularized linear regression) in DeepAutoQSAR hyper-parameter optimization [2022-4]
- New DeepAutoQSAR command line utility for greater ease-of-use [2022-4]
Desmond Molecular Dynamics
- In Trajectory Plots view Ramachandran plot of Protein Residues [2022-4]
Empirical and QM-based pKa Prediction
- Initial release of Epik 7, a new machine learning based application for fast pKa value and protonation state prediction [2022-4]
- Epik 7 can also produce a plot giving the populations of states as a function of pH
Solubility FEP (Beta)
- Option to show solubility results in logS unit [2022-4]
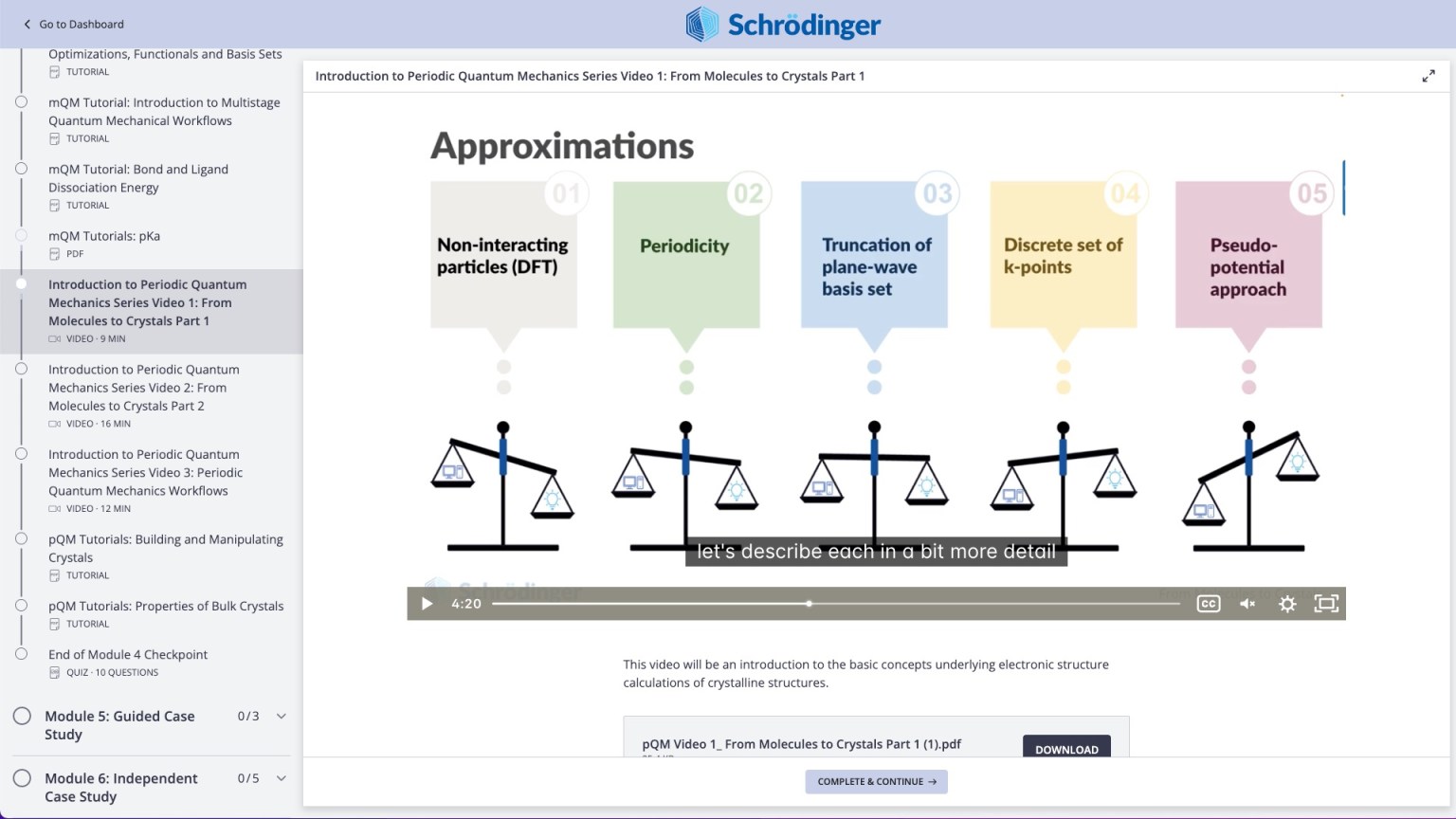
Quantum Mechanics
- Calculate ESP charges for excited states under the TDDFT/TDA approximation [2022-4]
- Complete calculations faster with parallel Jaguar calculations on Windows [2022-4]
- New Workflow Action Menu to guide to next steps for Plot Rigid/Relaxed scans [2022-4]
- Over 80 examples of Jaguar input files in documentation [2022-4]
Semi-Empirical Quantum Mechanics
- GFN2-xTB method now available in the Semiempirical Module panel [2022-4]
Biologics Drug Discovery
- To improve protein linker design, the loop database was updated to a new version specifically intended for interdomain linker design [2022-4]
- To expand chemical liability detection, Asp isomerization pattern and free cysteine detection were added to the Reactive Residues interface [2022-4]
- New command-line script for running Protein Interaction Analysis with the ability to export results to csv format [2022-4]
- First full release of Protein Descriptors interface which now supports .mae and .maegz files containing multiple structures [2022-4]
- Copy-paste sequences into Antibody Structure Prediction interface under new “Enter new sequence” option [2022-4]
- Reuse the same input csv file format for batch homology modeling in Antibody Structure Prediction when running from the command line or Maestro [2022-4]
- Added support for ‘keep glycan’ option in Antibody Structure Prediction during batch modeling [2022-4]
- Cysteine scanning panel for disulfide design now supports remote job submission which is useful for running large jobs e.g. using when MD trajectory as input [2022-4]
- Added “Antibody-Antigen” to Interactions scope dropdown [2022-4]
- Get Going with BioLuminate Video Series added to Documentation [2022-4]
Materials Science
GUI for Quantum ESPRESSO
- Quantum ESPRESSO GUI: Option to hide selected atoms [2022-4]
- Quantum ESPRESSO GUI: Upgraded NEB UI for improved UX [2022-4]
- Quantum ESPRESSO: Endpoints saved for NEB mae files at each iteration [2022-4]
- Quantum ESPRESSO: Reduced file size for custom saved NEB setups [2022-4]
- Quantum ESPRESSO: Use of automatic parallelization with GUI support [2022-4]
- Quantum ESPRESSO: -save_failures option for driver (command line) [2022-4]
- Quantum ESPRESSO: -last_only option for qe2mae tool to save the final structure only (command line) [2022-4]
- Quantum ESPRESSO: HUBBARD options enabled in input *.cfg (command line) [2022-4]
- Quantum ESPRESSO: Automatic restart for long AIMD simulations [2022-4]
- Quantum ESPRESSO: Support for cell volume relaxation [2022-4]
Molecular Dynamics
- Viscosity: ”,” used as the delimiter in CSV output for Einstein-Helfand analysis [2022-4]
Materials Informatics
- Machine Learning Property: Pre-built, validated machine learning models for a selective list of materials properties [2022-4]
- Molecular Descriptors: Report of semiempirical HOMO-LUMO gap [2022-4]
- Molecular Descriptors: Support for plotting molecular orbitals [2022-4]
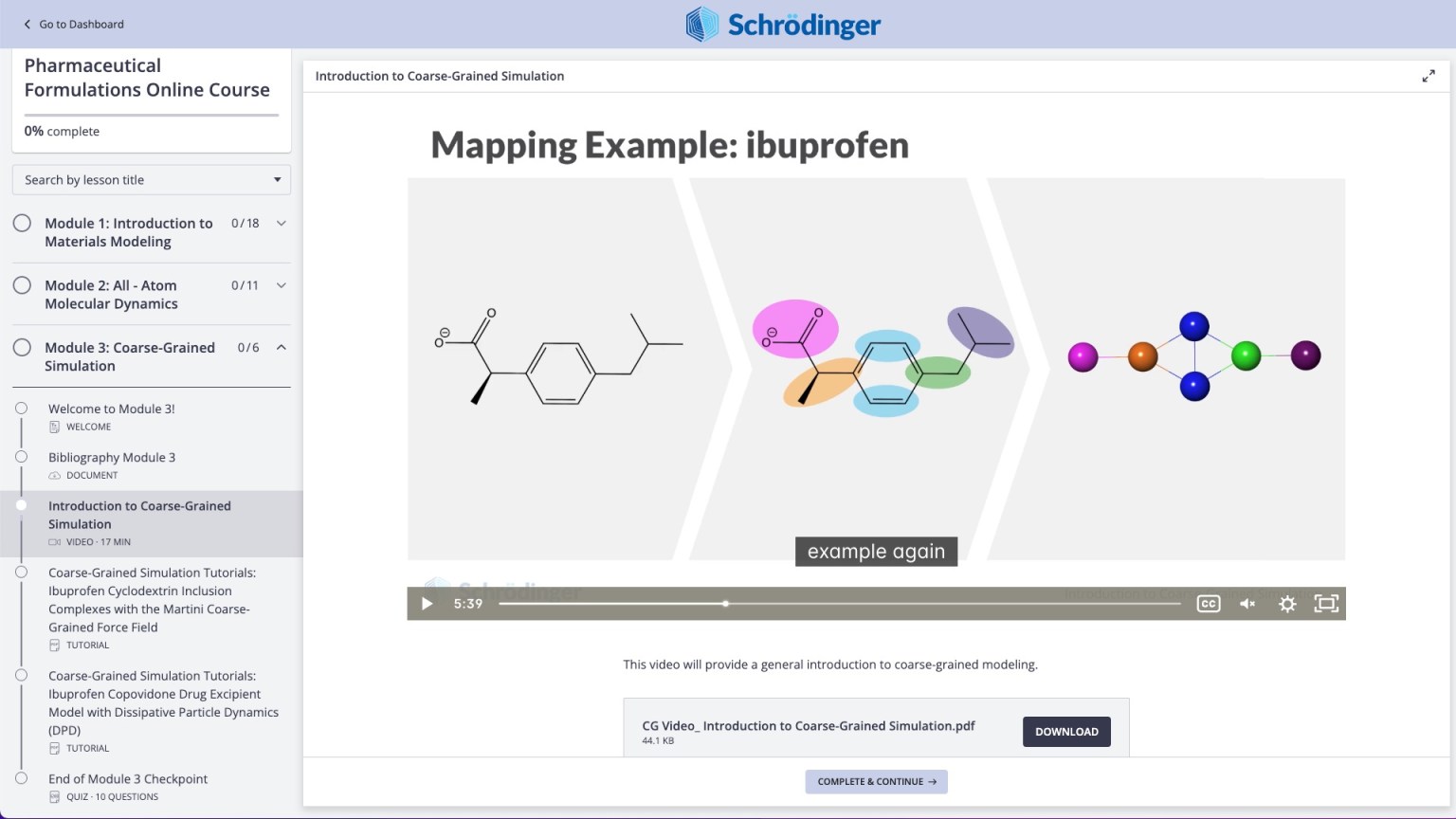
Coarse-Grained (CG) Molecular Dynamics
- Support for Ewald sums with Martini force field [2022-4]
- CG FF Builder: Option to export the viewer data to CSV [2022-4]
- CG FF Builder: Support for the use of existing trajectory [2022-4]
- Map Atoms to Particles: Option to map selected atoms from the input structure [2022-4]
Penetrant Loading Simulation
- Penetrant Loading: Robust handling of GCMC water models [2022-4]
- Penetrant Loading: Visualization of periodic unit cell for the output structures [2022-4]
MS Maestro User Interface
- Resized Trajectory Analysis task frame in Maestro for better user experience [2022-4]
MS Maestro Builders and Tools
- Complex Builder: Expansion of ligand library [2022-4]
- Disordered System: Option to generate cells with different numbers of molecules [2022-4]
- Elemental Enumeration: Jobs launched to queue instead of running interactively [2022-4]
- Manipulate Cell: Option to translate within -0.5 and 0.5 of the fractional coordinate [2022-4]
- Manipulate Cell: Support for change of lattice dimensions without FF retyping [2022-4]
- Query Bonds: Option to export output to CSV [2022-4]
- Semicrystalline Polymer: Option to use existing crystal (command line) [2022-4]
- Semicrystalline Polymer: Reporting percentage of crystallinity [2022-4]
- Semicrystalline Polymer: Speed-up for building with polymer models [2022-4]
Classical Mechanics
- MD Multistage: Improved estimation of timestep for Martini systems [2022-4]
- MD Multistage: Option to concatenate stages together for speed-up [2022-4]
- MD Multistage: Built-in Martini relaxation protocol suitable for NVT ensemble [2022-4]
- MD Multistage: Option to only write out selected molecules to trajectories (command line) [2022-4]
- Stress Strain: Output CSV updated at each new data point [2022-4]
- Thermophysical Properties: Support for Parrinello-Rahman barostat (command line) [2022-4]
Quantum Mechanics
- Band Shape: Option to add/select implicit solvent [2022-4]
- Organometallic Conformational Search: Option to select conformers after QM calculations (command line) [2022-4]
- Organometallic Conformational Search: Support for custom MacroModel COM files (command line) [2022-4]
- Organometallic Conformational Search: Support for MOPAC (command line) [2022-4]
Education Content
- Get Going with Materials Science Maestro Video Series added to Documentation [2022-4]
- Quick Reference Sheets available from both Documentation and Training webpages [2022-4]
- New Tutorial: Evaporation [2022-4]
- New Tutorial: Machine Learning Property Prediction [2022-4]
- Updated Tutorial: Polymer Electrolyte Analysis [2022-4]
- Updated Tutorial: Computing Atomic Charges [2022-4]
- Updated Tutorial: Activation Energies for Reactivity in Solids and on Surfaces [2022-4]
- Updated Tutorial: Organometallic Complexes [2022-4]
LiveDesign
What’s new in LiveDesign 2022-4
- The number of logins is enforced by a license limit: Users who attempt to log in after the number of available seats have been assigned will be denied access to LiveDesign
- Admins can forcibly log out users: Users can be forcibly logged out by removing their assigned Roles, or by specifying a specific username
- File Import Receipt: Receive feedback on file imports when the file contains errors, and instructions on how to correct the errors
- Sketcher Improvements:
- Implicit mode sketcher: Quickly switch between editing compounds and selecting a subset of the compound, by clicking directly on the select tools and draw tools
- Delete an atom by hovering over it and pressing the Backspace key on the keyboard
- Setting a protocol’s parameter to “Set Default” in the admin panel changes all existing models’ parameter to “Set Fixed”
- Interaction Surface within LigandDesigner: view the interaction surface to determine available growth space within a binding pocket
- Copying a single compound from the spreadsheet can be copied in a Mol v3000 format: a server wide setting permits copying a single molecule as either Extended SMILES or Mol v3000. Selecting and copying multiple compounds as once will copy the compounds using a SMILES format
- Freeform column picklist options can be reordered: Editing a Freeform column definition permits reordering the picklist options
- UI Improvements
- View more tiles on screen in Tile View, which has a much smaller tile size limit
- View more data in a spreadsheet cell; the “More Available…” message within spreadsheet cells has been replaced with a gradient to indicate additional data is in the cell
- Users can use Ctrl-Click to easily display results from additional 3D models in the 3D visualizer.
- Exported compound structure images will now include stereochemistry labels if these are turned on in the spreadsheet.
What’s Been Fixed
- Model columns can now be sorted and filtered when the cell contains both blank values and numeric or string values. Sorting will use the first non-empty value in the cell.
- Advanced searches with multiple Freeform column conditions return the same results even if the condition order is changed
- Compound images no longer show large atom labels when clicking on them within the main spreadsheet
- Project admins can edit all formulas within their projects
- Pinned plot tooltips in the visualize panel will reappear even after switching to another plot or another LiveReport
- Deleted LiveReports cannot be reopened by navigating directly to the LiveReport’s URL
- Pinned plot tooltips in forms update the connecting line when the view is resized
- Pasting multiple values into Filters will, once again, attempt to automatically identify the delimiter, or present an option to select the delimiter, by which to separate values.
- The LiveReport Manager dialog no longer obscures the last LiveReport with a horizontal scroll bar
- Bond angles for attachment points and carbon atoms from R-group decompositions are now displayed as angles of less than 180 degrees, while before they were displayed at a 180 degree angle
- LiveReports with 3D model returns no longer show a red error bar after opening
- Commons-text has been upgraded to patch security vulnerability CVE-2022-42889
- The 3D visualizer uses the high performance GPU on client computers to avoid crashes
- Formula results no longer disappear from the spreadsheet when columns used in the formula are hidden in the LiveReport
- The matched molecular pairs tool no longer fails to parse chiral compounds represented in an Extended SMILES format
- Error messages no longer sporadically appear when models are updated and saved in the Admin Panel
Training & Resources
Online Certification Courses
Level up your skill set with hands-on, online molecular modeling courses. These self-paced courses cover a range of scientific topics and include access to Schrödinger software and support.
Tutorials
Learn how to deploy the technology and best practices of Schrödinger software for your project success. Find training resources, tutorials, quick start guides, videos, and more.















Trusted by global leaders in industry